Portfolio
Selected Projects
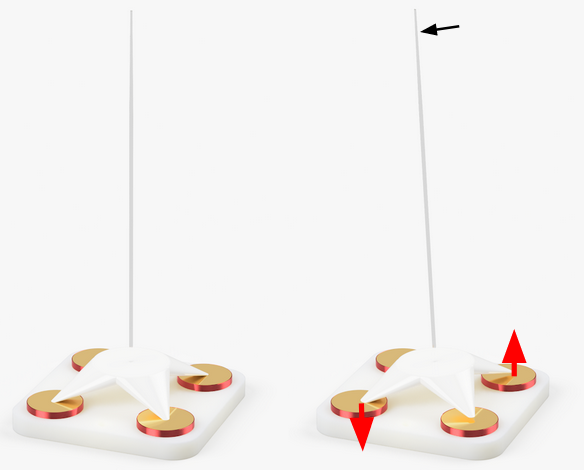
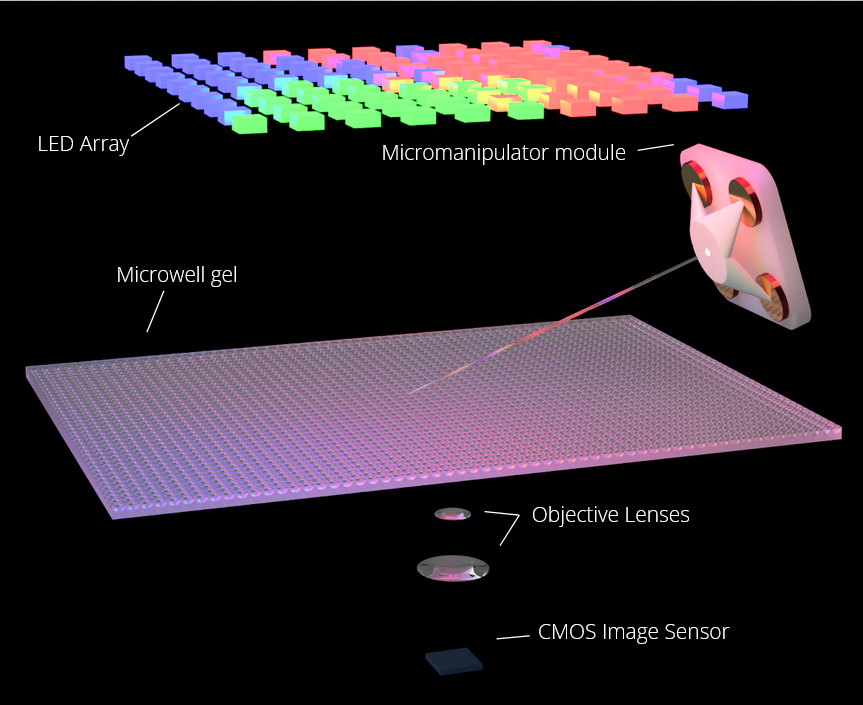
3-D Localization of Micromanipulators Using Microscopy for Autonomous Visual Servoing (In-Progress)
Presented at Optica Imaging and Applied Optics Congress 2022
We developed a method to rapidly determine the 3-D position of a glass-pipette micromanipulator using color-coded illumination and optical microscopy. This technology has potential to enable autonomous visual servoing and multi-manipulator systems for highly parallelized cell manipulation.
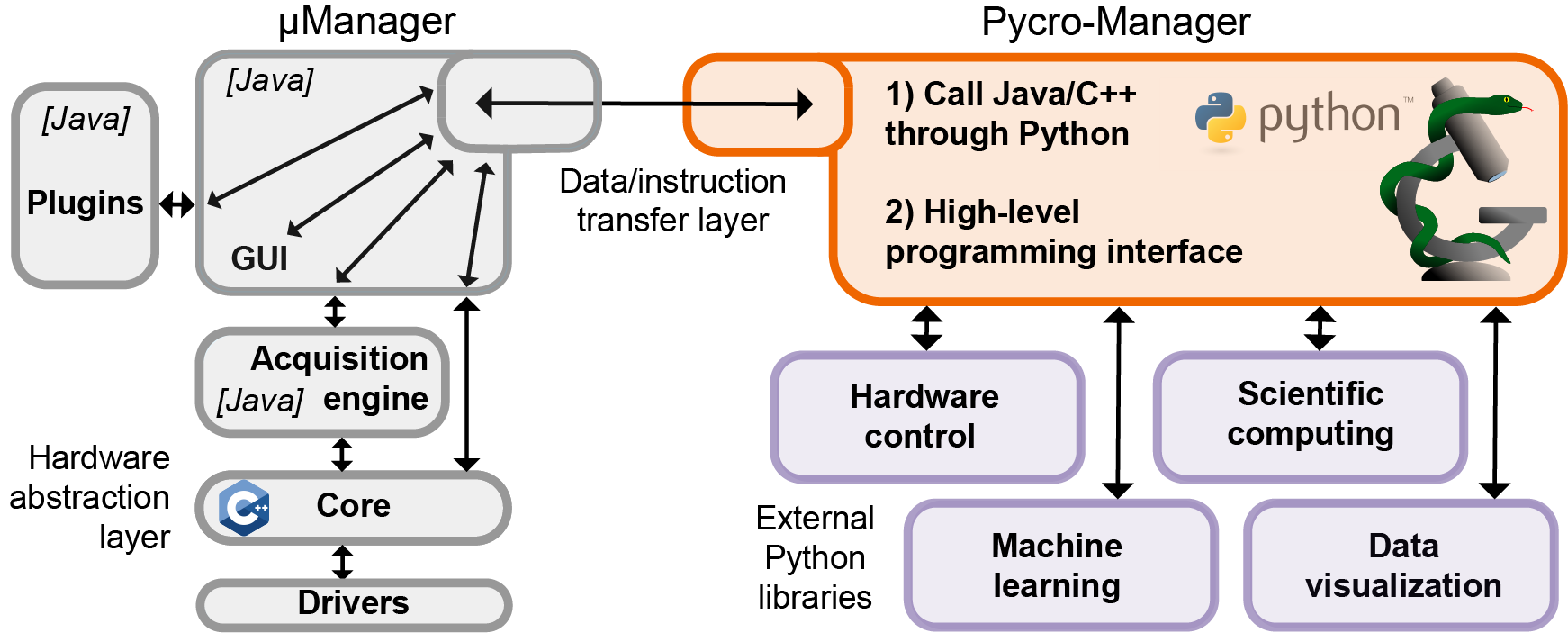
Pycro-Manager: open-source software for customized and reproducible microscope control
Nature Methods Article Documentation
Pycro-Manager is a multi-purpose python package for the control of microscope hardware and the acquisition of data. It is designed for performance and flexibility to be used in a variety of applications, ranging from automating data collection across multiple microscopes for high-throughput biology, to enabling easier control of microscopes in academic labs.
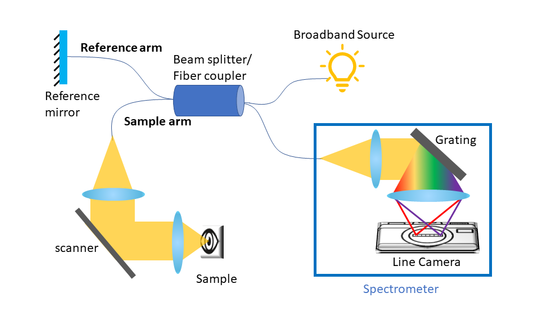
Compact Optical Coherence Tomography with a Broadband Superluminescent Diode (In-Progress)
We are currently working on fabricating a spectral domain OCT system for retinal images that utilizes broadband SLD. The focus of the project is to engineer a portable and affordable eye imaging device.
Collaborators: Guanghang Meng
APRS SDR + Video Compression + Healing Compression Artifacts Using ML
The goal of this project was to efficiently transmit video data over APRS. We implemented an FM transmitter/receiver and APRS TNC from scratch on a software-defined radio. To reduce the number of data packets required, we implemented a compression algorithm based on H.264, utilizing DCT compression, chroma downsampling, motion prediction, zero run-length encoding, and LZMA. To eliminate compression artifacts, we designed a ResNet to predict the ground truth frames of a video from compressed frames. We created a training dataset with frames from 78,000 videos from the Vimeo90k dataset, and trained the network over 24 hours. Our algorithm was able to acheive a 61x compression ratio on 328kB video while maintaining a 27dB PSNR.
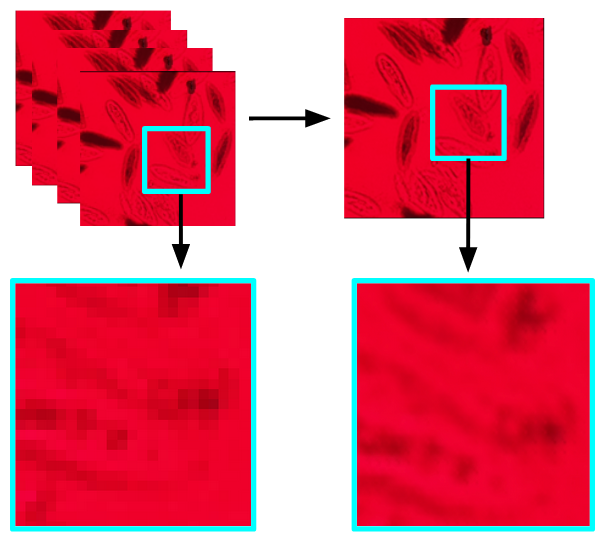
Scoplet: Compact Holographic Computational Microscopy
In this project we created a portable digital in-line holographic microscope. The optical path is extraordinarily simple: just a single led and a sample slide placed ~0.5 mm in front of an image sensor. In order to improve the resolution of the diffraction patter we measure, we implemented a super resolution algorithm that leverages multiple images of the subject with random non-integer shifts applied to the sensor between images via a small buzzer, and we achieved up to a 2x improvement in resolution. We computational backpropagate the diffraction pattern and algorithmically “refocus” the diffraction pattern into an image.
Collaborators: Leyla Kabuli, Franklin Wang
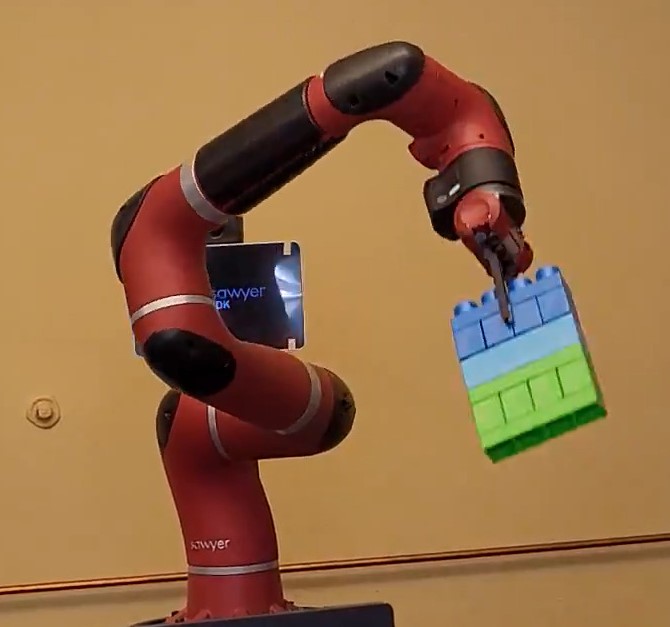
Visual Servoing for Lego Manipulation
We used a Rethink Robotics Sawyer robotic arm to autonomously locate lego blocks using the in-arm camera and manipulate the blocks. We created a forward/inverse kinematics engine from scratch, implemented a basic occupancy grid tracker, trained a segmentation algorithm, and created a closed loop controller to position the arm.
Collaborators: Will Panitch
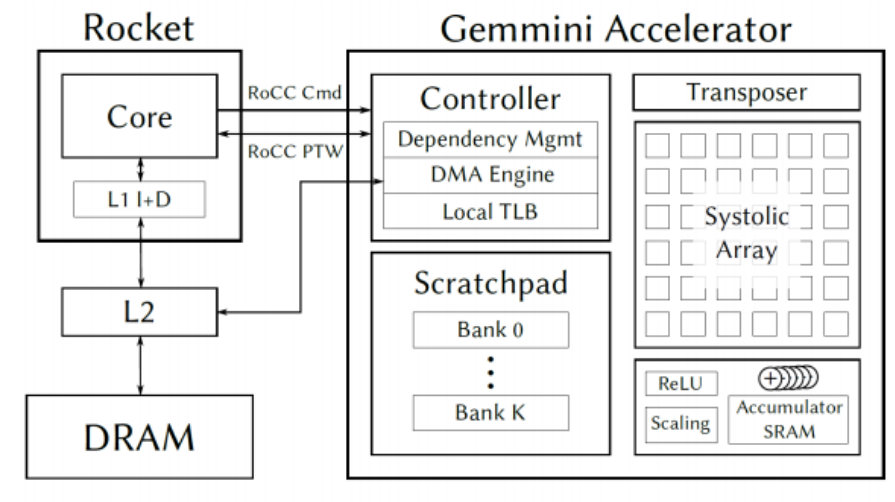
RISC V CPU with Neural Coprocessor
We implemented a complete 3-stage pipelined RISC V CPU from scratch in Verilog, including a branch predictor, forwarding logic, caches, memory mapped I/Os (PWM, UART, SPI, and audio interfaces). To test the I/O we wrote a piano program in assembly that allowed the user to play tones by pressing keys on another computer communicating with our CPU over UART. We created a FP16 systolic array matrix-multiplication coprocessor from scratch to accelerate inference for neural networks. The CPU core and accelerator were integrated into an SoC using the Rocket Chip framework. Finally, the entire system was put in hardware on a Xilinx Zynq UltraScale+ FPGA, and we demonstrated hardware acceleration of a simple speech-to-text neural network.
Collaborators: Oskar Hurst
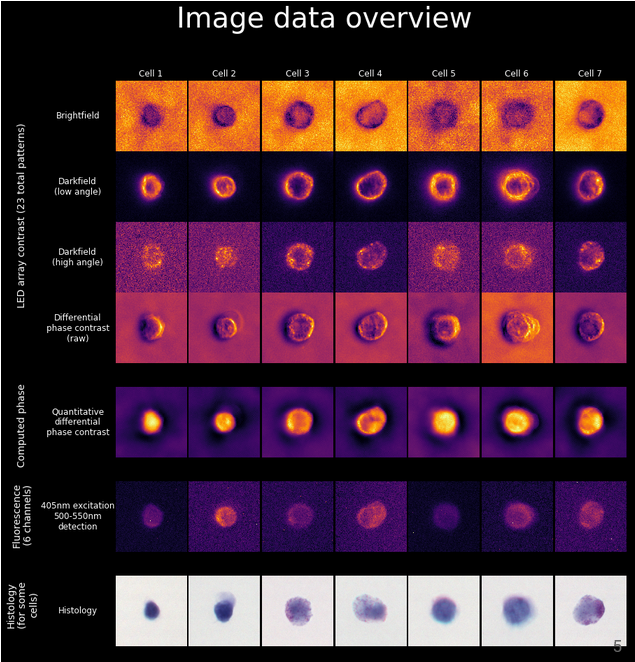
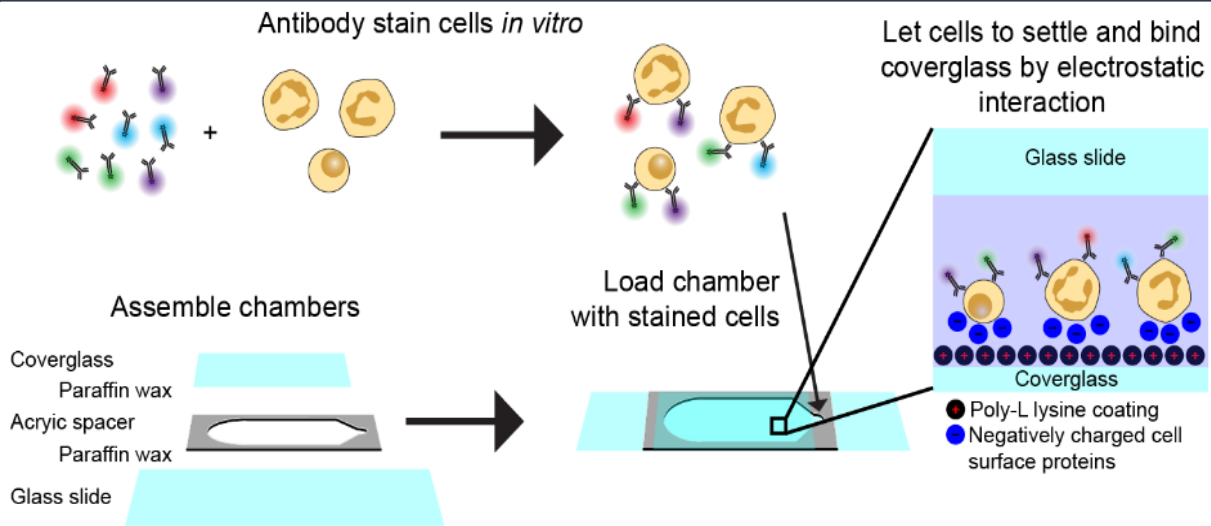
Berkeley Single Blood Cell Microscopy (BSBCM) Dataset: Unsupervised Analysis of Cellular Morphology Using VQ-VAEs
We collected microscopy images of 412,000 individual blood cells using coded illumination to reconstruct label-free brightfield, darkfield, and quantitative phase contrast images of the cells. We also imaged the cells with 6 fluorescent antibody probes labeling major cellular components (membrane, nucleus, mitchondria, etc.), and brightfield images of histology stains.
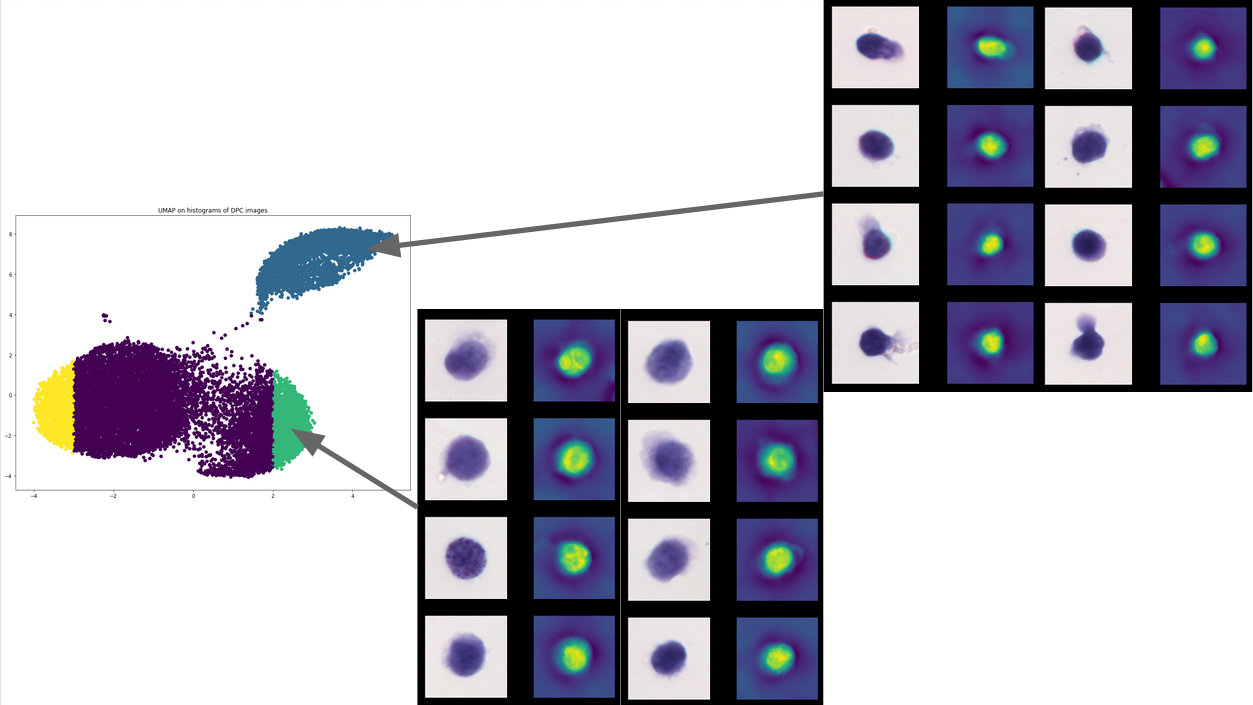
To investigate the information overlap between the label-free microscopy methods and the chemically labeled (fluorescent and histology) methods, we trained a vector-quantized variational autoencoder (VQ-VAE) to try and predict the labeled images using the label-free images as input. We found that quantitative phase is especially rich in morphological information.
Sampling the latent space of the VQ-VAE and performing a UMAP on the output for the entire dataset, we found that the encoder was effectively encoding information about the cell type of the different blood cells in the dataset.
Collaborators: Henry Pinkard
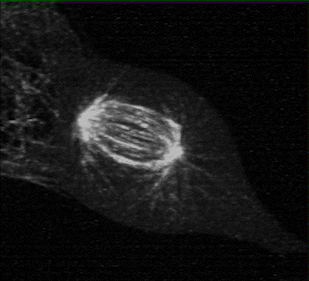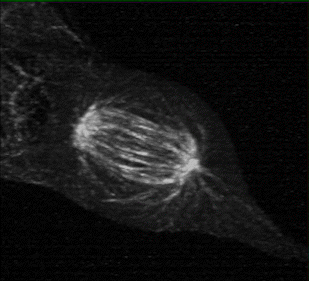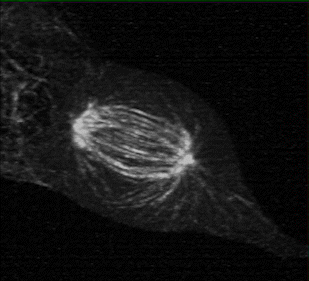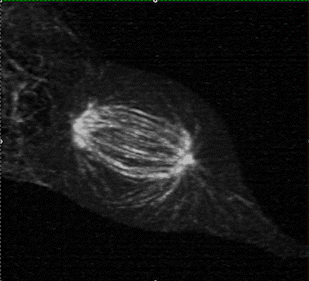
Measuring K-fiber and Interpolar Microtubule Chirality Throughout Mitosis
The mitotic spindle is an incredibly complex mechanism composed of a self-assembling molecular structure that physically separates chromosomes to the two daughter cells during mitosis. This structure of microtubules, microtubules associated proteins (motors, crosslinker, etc.), and the kinetochore, enables the persistence of eukaryotic life on Earth.
Using 3D lattice light sheet microscopy in the MOSAIC at the Advanced Bioimaging Center at UC Berkeley, we were one of the first researchers to characterize the presence of chirality, or “twist” in the fibers of the mitotic spindle, during cell division.
Collaborators: Caleb Rux, Brian Blankenship
Colorizing the Prokudin-Gorskii Photo Collection. Reconstructing some of the first ever color photographs.
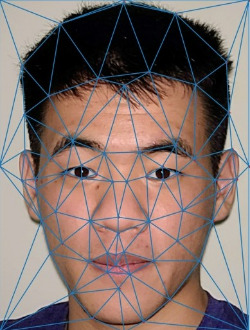
Face morphing. Exploring triangulation and affine transforms.





 https://photos.app.goo.gl/qLHeagTy9eL9qM968
https://photos.app.goo.gl/qLHeagTy9eL9qM968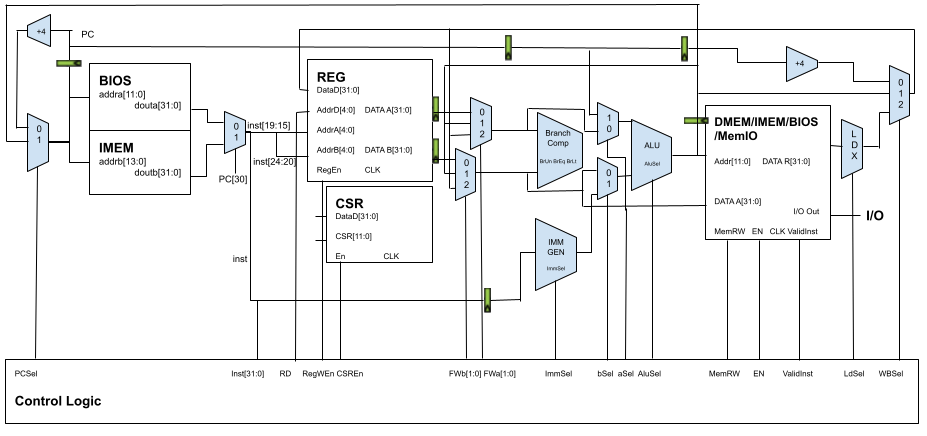
.gif)